Single Particle Analysis Simulations
- Single Particle Analysis Schematic
- STEM Simulations
- Single Slice Reconstruction
- Reconstructs 3D volumes of homogeneous samples from a large number of 2D EM randomly-oriented micrographs
- The main steps involve:
- Obtaining cryo-EM micrographs
- Automated "particle" identification
- Class averaging
- Orientation estimation
- Tomographic reconstruction

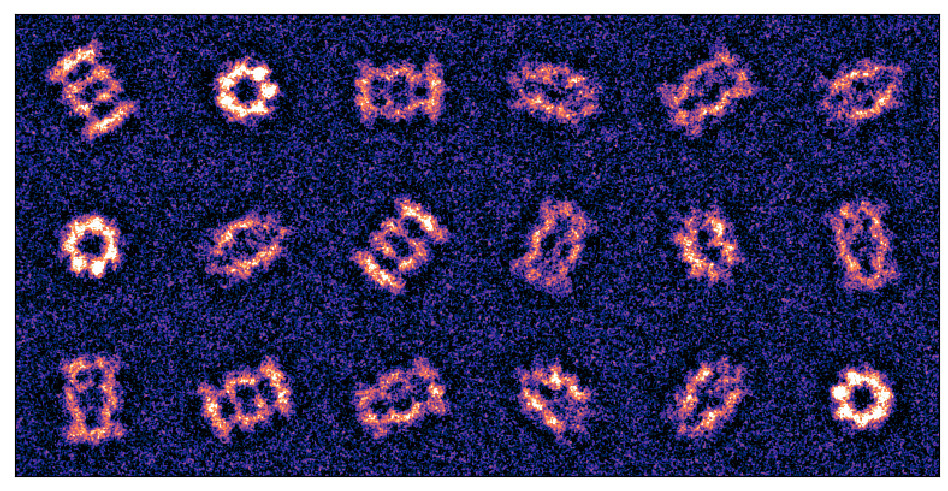
- T20S Proteasome
- Projected Potential
- Noisy Diffraction Patterns
- We use a solved structure, namely the T20S proteasome
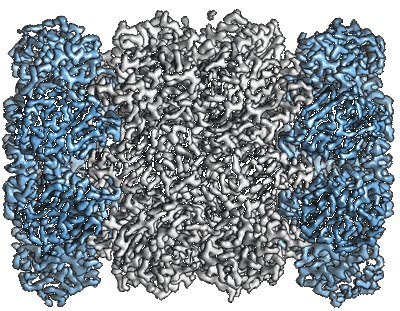
- We use a solved structure, namely the T20S proteasome
- We simulate a large FOV using 16x16 tiled proteasome volumes, each randomly rotated about its center
- 300 kV, 2.5μm defocus, 3 mrad convergence angle
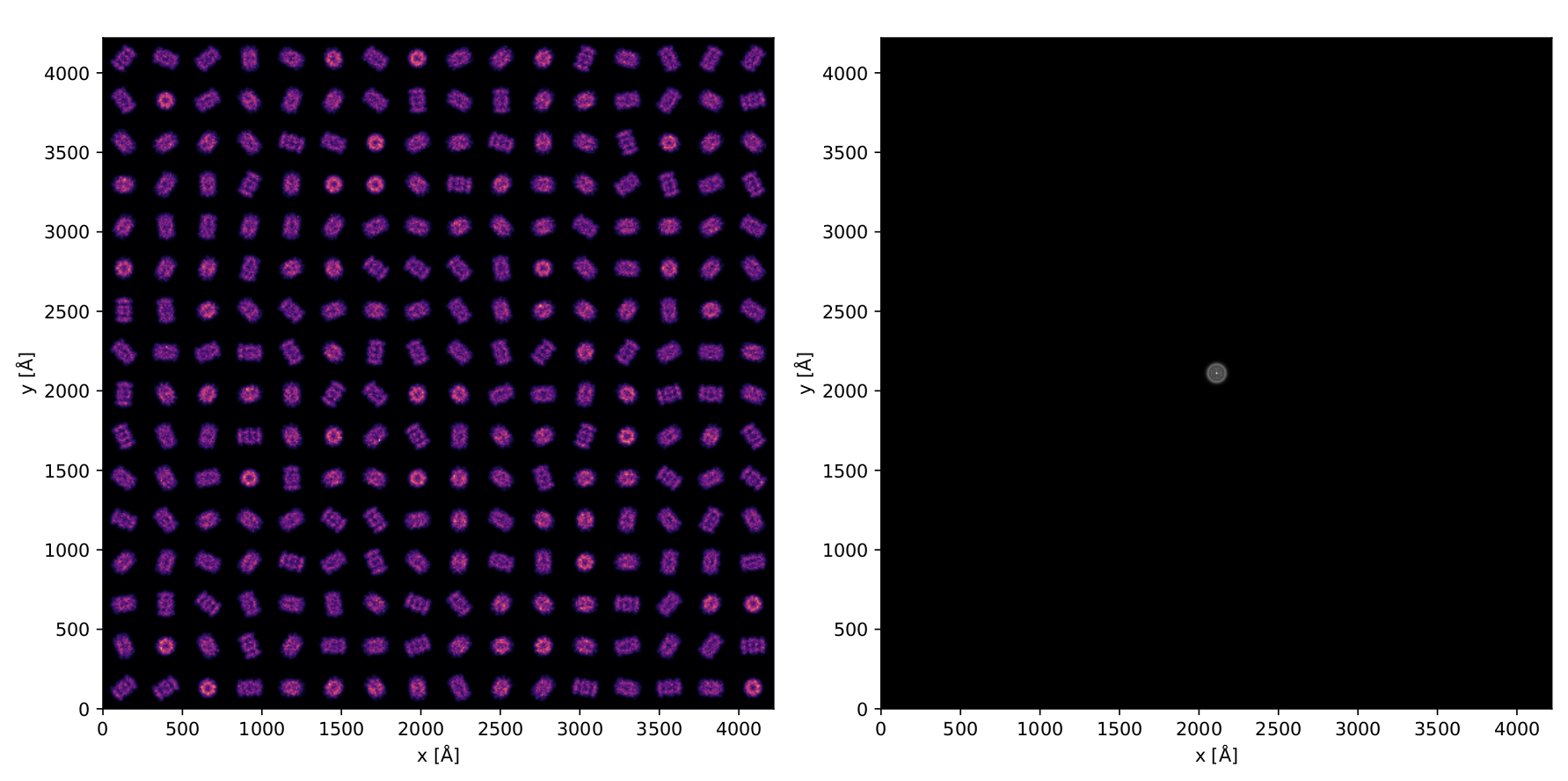
- We use a solved structure, namely the T20S proteasome
- We simulate a large FOV using 16x16 tiled proteasome volumes, each randomly rotated about its center
- 300 kV, 2.5μm defocus, 3 mrad convergence angle
- We use a finite dose of 30 e/Å2
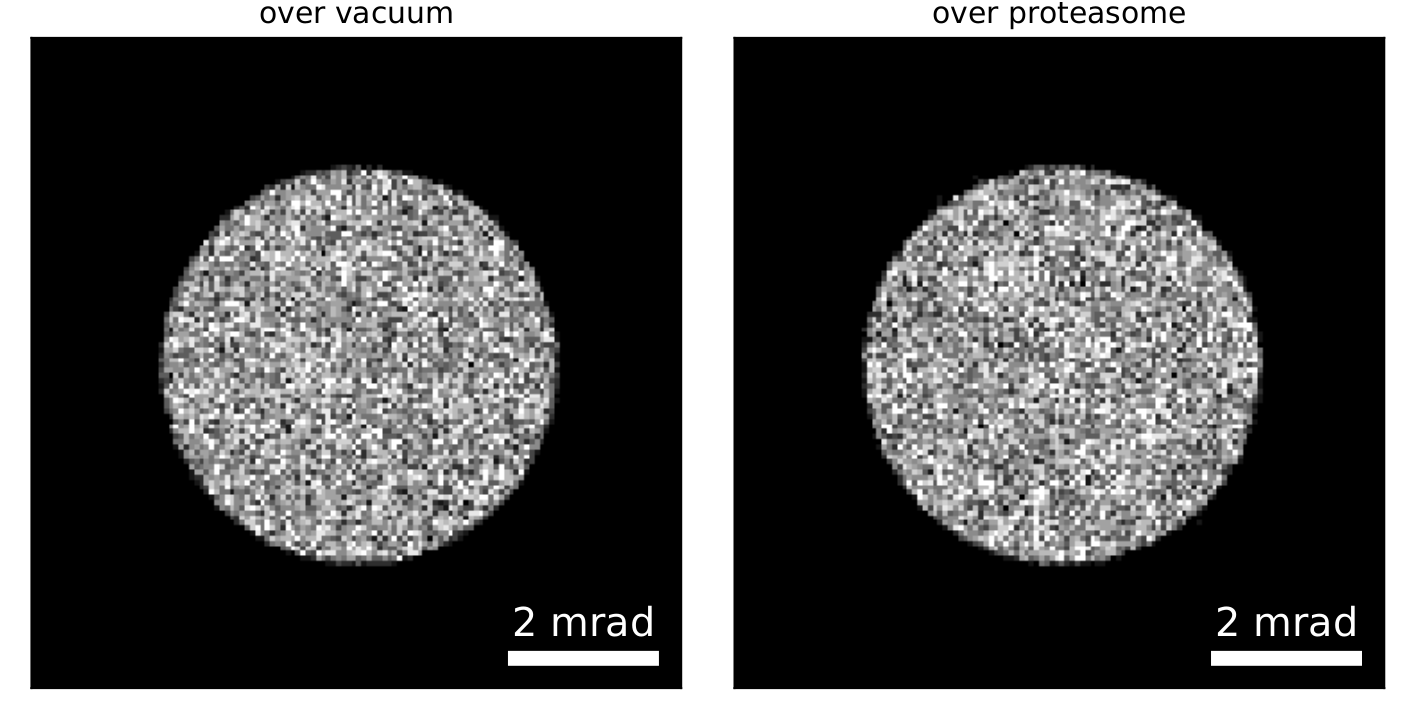
- We start by reconstructing the entire FOV using single-slice ptychography